Biophysics
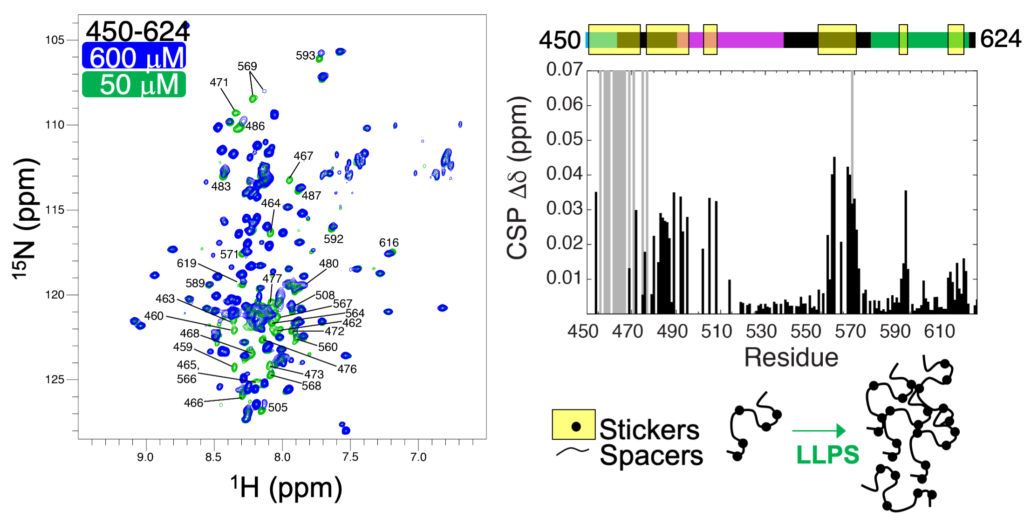
We have used NMR spectroscopy to probe the structure and dynamics of proteins on an atomic level. 15N-1H HSQC spectra, like the one shown below (left), probe the microenvironment of backbone amide resonances (N-H). Using the C-terminal construct of UBQLN2, residues 450-624, we determined that residues 450-580 are largely intrinsically-disordered. We used NMR to probe the concentration dependence of UBQLN2 450-624 amide chemical shifts, and quantified these as CSPs (chemical shift perturbations). We found that many resonances disappeared as concentration increased (gray bars in CSP plot) or changed position, consistent with UBQLN2’s ability to oligomerize. We hypothesized that these residues “stickers”, the parts of the protein that are involved in UBQLN2 self-association and in driving phase separation. The other residues are “spacers”, segments that connect “stickers” together.
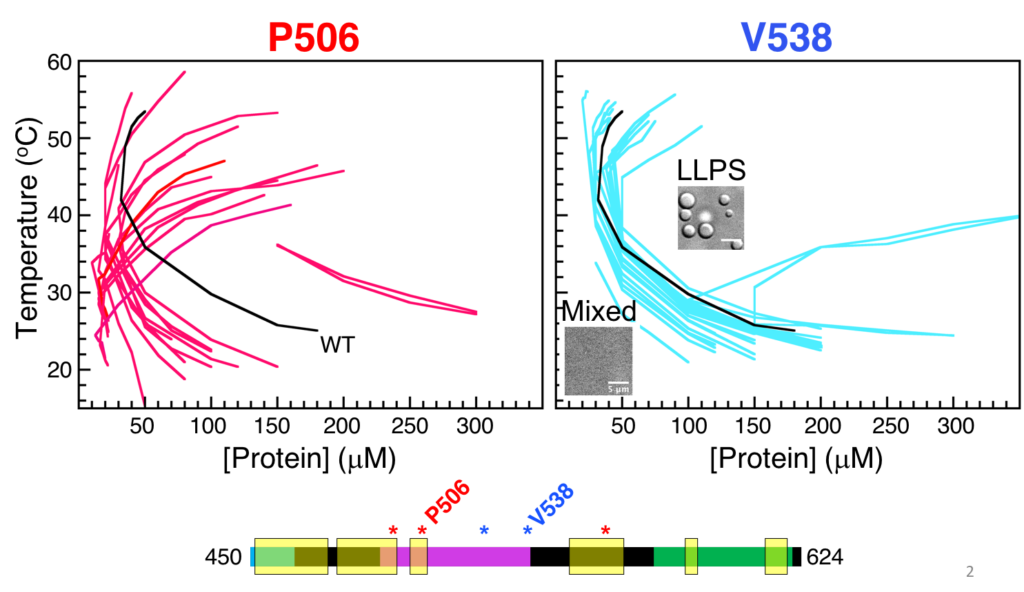
To test whether residues are “stickers” or “spacers”, we decided to screen the effects of amino acid substitutions on UBQLN2 phase separation at representative “sticker” and “spacer” positions. Using the CSP plot above, we chose three sticker positions (497, 506, and 564), and two spacer positions (525 and 538). We generated a total of 95 UBQLN2 mutants (substituting each of the natural amino acids at each of the positions one at a time), and screened phase separation using turbidity assays.
For two of the positions, we obtained the low concentration arm of temperature-concentration phase diagrams (see below). Strikingly, substitutions at the spacer position did not change the overall shape of the UBQLN2 phase diagram (except for acidic residues), whereas sticker substitutions dramatically shifted the shape and position of the UBQLN2 phase diagram. Some of this work is recently published (Yang et al., J Phys Chem B, 2019).
It is important to note that the “sticker” positions coincide with some of the known disease-linked substitution sites. As a next step, we are currently investigating the effects of “sticker” disease-linked mutations on UBQLN2 phase separation.
We find that the “stickers” and “spacers” framework is a very useful formalism for determining the driving forces of phase separation behavior of biopolymers. See some of the excellent work and reviews in this field (Semenov and Rubinstein, 1998; Choi et al., 2019; Choi, Holehouse, Pappu, 2019).